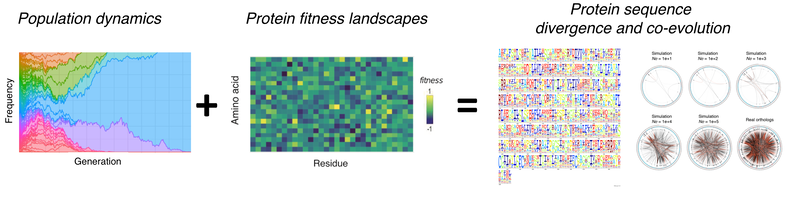
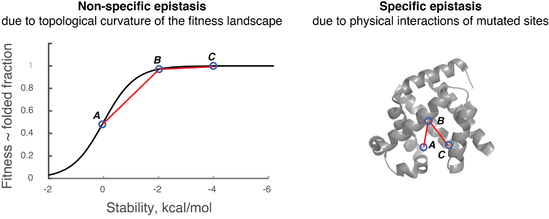
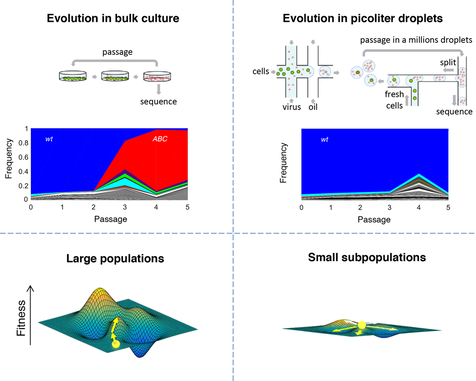
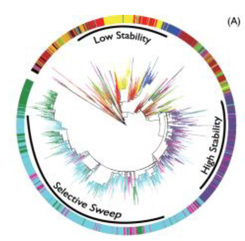
|
*Corresponding author
#Equal contribution |
2024
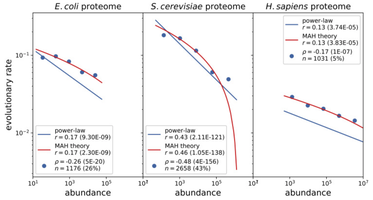
45. Genetic landscape of an in vivo protein interactome
Savandara Besse, Tatsuya Sakaguchi, Louis Gauthier, Zahra Sahaf, Olivier Péloquin, Lidice Gonzalez, Xavier Castellanos-Girouard, Nazli Koçatug, Chloé Matta, Julie G. Hussin, Stephen W. Michnick, Adrian W.R. Serohijos*
2023 In revision
Biorxiv
Savandara Besse, Tatsuya Sakaguchi, Louis Gauthier, Zahra Sahaf, Olivier Péloquin, Lidice Gonzalez, Xavier Castellanos-Girouard, Nazli Koçatug, Chloé Matta, Julie G. Hussin, Stephen W. Michnick, Adrian W.R. Serohijos*
2023 In revision
Biorxiv
44. Intra- and inter-species interactions drive early phases of invasion in mice gut microbiota
Gencel M, Cofino GM, Hui C, Sahaf Z, Gauthier L, Tsang D, Philpott D, Ramathan S, Menendez A, Bershtein S, Serohijos AWR*
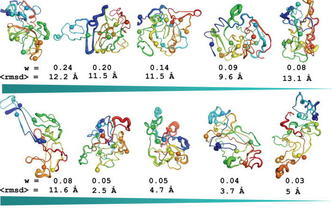
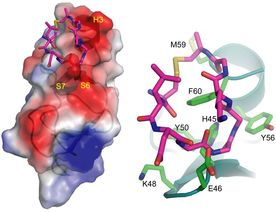
(2023) In revision.
Biorxiv
Gencel M, Cofino GM, Hui C, Sahaf Z, Gauthier L, Tsang D, Philpott D, Ramathan S, Menendez A, Bershtein S, Serohijos AWR*
(2023) In revision.
Biorxiv
43. Non-consecutive enzyme interactions within TCA cycle supramolecular assembly regulate carbon-nitrogen metabolism
Weronika Jasinska, Mirco Dindo, Sandra M. Correa, Adrian W.R. Serohijos, Paola Laurino*, Yariv Brotman*, Shimon Bershtein*
2023 Under review.
Biorxiv
Weronika Jasinska, Mirco Dindo, Sandra M. Correa, Adrian W.R. Serohijos, Paola Laurino*, Yariv Brotman*, Shimon Bershtein*
2023 Under review.
Biorxiv
42. Neutral Drift and Threshold Selection Promote Phenotypic Variation
Ayşe N. Erdoğan, Pouria Dasmeh, Raymond D. Socha, John Z. Chen, Ben Life, Rachel Jun, Linda Kiritchkov, Dan Kehila, Adrian W.R. Serohijos, Nobuhiko Tokuriki
(2023) In revision.
Biorxiv
Ayşe N. Erdoğan, Pouria Dasmeh, Raymond D. Socha, John Z. Chen, Ben Life, Rachel Jun, Linda Kiritchkov, Dan Kehila, Adrian W.R. Serohijos, Nobuhiko Tokuriki
(2023) In revision.
Biorxiv
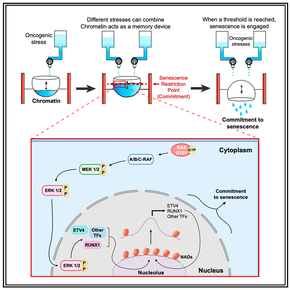
41. A senescence restriction point acting on chromatin integrates oncogenic signals.
Lopes-Paciencia S, Bourdeau V, Rowell MC, Amirimehr D, Guillon J, Kalegari P, Barua A, Quoc-Huy Trinh V, Azzi F, Turcotte S, Serohijos A, Ferbeyre G.
Cell Reports 2024 Apr 2;43(4):114044. doi: 10.1016/j.celrep.2024.114044. Online ahead of print.
(PDF)
Lopes-Paciencia S, Bourdeau V, Rowell MC, Amirimehr D, Guillon J, Kalegari P, Barua A, Quoc-Huy Trinh V, Azzi F, Turcotte S, Serohijos A, Ferbeyre G.
Cell Reports 2024 Apr 2;43(4):114044. doi: 10.1016/j.celrep.2024.114044. Online ahead of print.
(PDF)
2023
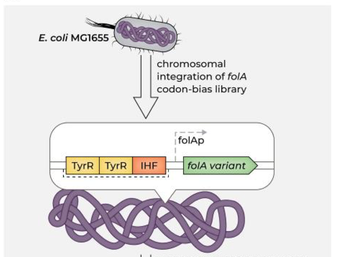
40. The Fitness Effects of Codon Composition of the Horizontally Transferred Antibiotic Resistance Genes Intensify at Sub-lethal Antibiotic Levels.
Shaferman M, Gencel M, Alon N, Alasad K, Rotblat B, Serohijos AWR, Alfonta L, Bershtein S.
Mol Biol Evol. 2023 Jun 1;40(6):msad123. doi: 10.1093/molbev/msad123.
(PDF)
Shaferman M, Gencel M, Alon N, Alasad K, Rotblat B, Serohijos AWR, Alfonta L, Bershtein S.
Mol Biol Evol. 2023 Jun 1;40(6):msad123. doi: 10.1093/molbev/msad123.
(PDF)
2021
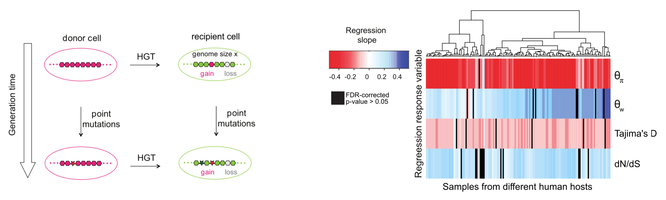
39. Mobile gene sequence evolution within individual human gut microbiomes is better explained by gene-specific than host-specific selective pressures.
N'Guessan A, Brito IL, Serohijos AWR*, Shapiro BJ*
(2021) Genome Biol Evol., in press doi: 10.1093/gbe/evab142.
N'Guessan A, Brito IL, Serohijos AWR*, Shapiro BJ*
(2021) Genome Biol Evol., in press doi: 10.1093/gbe/evab142.
38. Avoidance of protein unfolding constrains protein stability in long-term evolution
Razban RM, Dasmeh P, Serohijos AWR, Shakhnovich EI*
(2021) Biophys J. 120(12):2413-2424. doi: 10.1016/j.bpj.2021.03.042.
Razban RM, Dasmeh P, Serohijos AWR, Shakhnovich EI*
(2021) Biophys J. 120(12):2413-2424. doi: 10.1016/j.bpj.2021.03.042.
2020
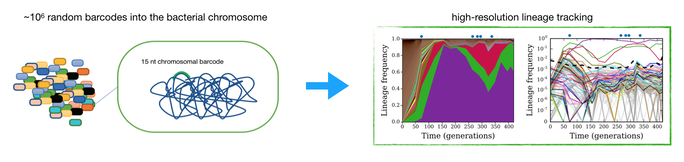
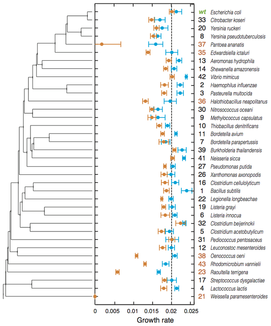
37. Chromosomal barcoding of E. coli populations reveals lineage diversity dynamics at high resolution
J Lerner, M Manhart, W Jasinska, L Gauthier, A Serohijos, S Bershtein*
(2020) Nature Eco & Evol. doi: 10.1038/s41559-020-1103-z. (PDF)
J Lerner, M Manhart, W Jasinska, L Gauthier, A Serohijos, S Bershtein*
(2020) Nature Eco & Evol. doi: 10.1038/s41559-020-1103-z. (PDF)
36. L Gauthier, Stynen B, A Serohijos*, SW Michnick*
Genetics’ piece of the PI: Inferring the origin of complex traits and diseases from proteome-wide protein-protein interaction dynamics
(2020) Bioessays , 42(2):e1900169. (PDF)
Genetics’ piece of the PI: Inferring the origin of complex traits and diseases from proteome-wide protein-protein interaction dynamics
(2020) Bioessays , 42(2):e1900169. (PDF)
2019
35. M Mariani , P Dasmeh , A Fortin , E Caron , M Kalamujic , AN Harrison , DM Kasumba , S Cervantes-Ortiz , E Mukawera , A Serohijos , N Grandvaux*
The Combination of IFN β and TNF Induces an Antiviral and Immunoregulatory Program via Non-Canonical Pathways Involving STAT2 and IRF9
(2019) Cells 8(8). pii: E919. (PDF)
The Combination of IFN β and TNF Induces an Antiviral and Immunoregulatory Program via Non-Canonical Pathways Involving STAT2 and IRF9
(2019) Cells 8(8). pii: E919. (PDF)
34. S Khoury, M Piltonen, A-T Ton, T Cole, A Samoshkin, S Smith, I Belfer, G Slade, R Fillingim, J Greenspan, R Ohrbach, W Maixner, G Neely, A Serohijos, L Diatchenko*
A functional missense substitution in Dopa Decarboxylase worsens somatic symptoms through the serotonin pathway
(2019) Ann Neurol. 86(2):168-180. (PDF)
A functional missense substitution in Dopa Decarboxylase worsens somatic symptoms through the serotonin pathway
(2019) Ann Neurol. 86(2):168-180. (PDF)
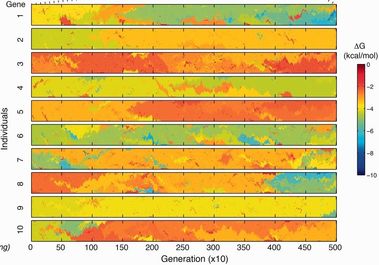
33. L Gauthier, R Di Franco, A Serohijos*,
SodaPop: A computational suite for simulating the dynamics of asexual populations on protein fitness landscapes
(2019) Bioinformatics 35 (20), 4053-4062
SodaPop: A computational suite for simulating the dynamics of asexual populations on protein fitness landscapes
(2019) Bioinformatics 35 (20), 4053-4062
2018
32. P Dasmeh, A Serohijos*
Estimating the contribution of folding stability to non-specific epistasis in protein evolution
(2018) Proteins. 86(12):1242-1250. doi: 10.1002/prot.25588.
Estimating the contribution of folding stability to non-specific epistasis in protein evolution
(2018) Proteins. 86(12):1242-1250. doi: 10.1002/prot.25588.
31. A Rotem#, A Serohijos#, C Chang, J Wolfe, A Fischer, T Mehoke, H Zhang, Y Tao, L Ung, J-M Choi, A Kolawole, S Koehler, S Wu, P Thielen, N Cui, P Demirev, N Giacobbi, T Julian, K Schwab, J Lin, T Smith, J Pipas, C Wobus, A Feldman, D Weitz*, E Shakhnovich*
“Evolution on the biophysical fitness landscape of an RNA virus”
(2018) Mol Biol Evol. , 35(10):2390-2400. doi: 10.1093/molbev/msy131.
30. E Klein, D Blumenkrantz, A Serohijos, E Shakhnovich, J-M Choi, J Rodrigues, B Smith, A Lane, A Feldman, A Pekosz*,
“Stability of the influenza hemagglutinin protein correlates with fitness and seasonal evolutionary dynamics"
(2018) mSphere 3:e00554-17. doi.org/10.1128/mSphereDirect.00554-17
2017
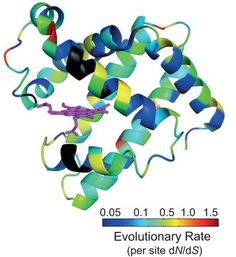
29. P. Dasmeh#, E. Girard#, A Serohijos*
"Highly expressed genes evolve under strong epistasis from a proteome-wide scan in E. coli"
(2017) Scientific Reports 7: 15844 | DOI:10.1038/s41598-017-16030-z . (PDF)
"Highly expressed genes evolve under strong epistasis from a proteome-wide scan in E. coli"
(2017) Scientific Reports 7: 15844 | DOI:10.1038/s41598-017-16030-z . (PDF)
28. G Desrochers, L Cappadocia, M Lussier-Price, A-T Ton, R Ayoubi, A Serohijos, JG Omichinski, A Angers*,
"Molecular Basis of Interactions Between SH3 Domain-Containing Proteins and the Proline-Rich Region of the Ubiquitin Ligase Itch"
(2017) J. Biol. Chem. 292(15):6325-6338. (PDF)
"Molecular Basis of Interactions Between SH3 Domain-Containing Proteins and the Proline-Rich Region of the Ubiquitin Ligase Itch"
(2017) J. Biol. Chem. 292(15):6325-6338. (PDF)
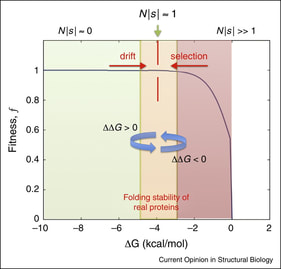
27. S Bershtein, A Serohijos, EI Shakhnovich*,
"Bridging the physical scales in evolutionary biology: From protein sequence space to fitness of organisms and populations"
(2017) Curr. Opin. Struct. Biol. 42: 31-40. (PDF)
"Bridging the physical scales in evolutionary biology: From protein sequence space to fitness of organisms and populations"
(2017) Curr. Opin. Struct. Biol. 42: 31-40. (PDF)
2016
26. N Cheron#, A Serohijos#, D Lucarelli, S Murphy, J Lin, AB Feldman and EI Shakhnovich,
“Evolutionary dynamics of viral escape under antibody stress: A biophysical model”
(2016) Protein Sci. doi: 10.1002/pro.2915. [#Equal contribution] (PDF)
“Evolutionary dynamics of viral escape under antibody stress: A biophysical model”
(2016) Protein Sci. doi: 10.1002/pro.2915. [#Equal contribution] (PDF)
2015
25. S Bershtein#, A Serohijos#, S Bhattacharyya, M Manhart, J-M Choi, W. Mu, J Zhou, EI Shakhnovich,
"Protein homeostasis imposes a barrier to the functional integration of horizontally transferred genes in bacteria"
(2015) PLoS Genetics 11(10): e1005612. [#Equal contribution] (PDF)
"Protein homeostasis imposes a barrier to the functional integration of horizontally transferred genes in bacteria"
(2015) PLoS Genetics 11(10): e1005612. [#Equal contribution] (PDF)
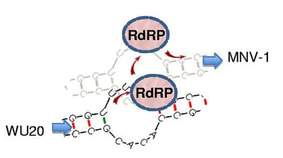
24. H Zhang, SK Cockrell, AO Kolawole, Assaf Rotem, A Serohijos, TS Mehoke, JS Lin, NS Giacobbi, EI Shakhnovich, AB Feldman, DA Weitz, CE Wobus, and JM Pipas
“Isolation and analysis of rare norovirus recombinants from co-infected mice using droplet-based microfluidics”
(2015) J Virology 89(15):7722-34. (PDF)
“Isolation and analysis of rare norovirus recombinants from co-infected mice using droplet-based microfluidics”
(2015) J Virology 89(15):7722-34. (PDF)
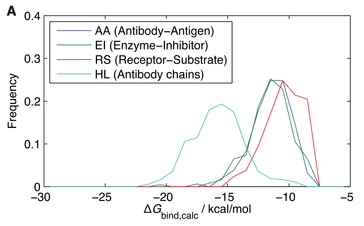
23. J.M. Choi, A Serohijos, S. Murphy, D. Lucarelli, L. Lofranco, A.B. Feldman and E.I. Shakhnovich,
“Minimalistic Predictor of Protein Binding Energy: Contribution of Solvation Factor to Protein Binding”
(2015) Biophys J. 108(4):795-8. (PDF)
“Minimalistic Predictor of Protein Binding Energy: Contribution of Solvation Factor to Protein Binding”
(2015) Biophys J. 108(4):795-8. (PDF)
2014
22. P. Dasmeh*,#, A Serohijos*,#, K. Kepp, E.I. Shakhnovich,
"The influence of selection for folding stability on dN/dS estimations"
(2014), Genome Biology and Evolution. 6 (10): 2956-2967. [*Equal contribution; #Corresponding authors] (PDF)
"The influence of selection for folding stability on dN/dS estimations"
(2014), Genome Biology and Evolution. 6 (10): 2956-2967. [*Equal contribution; #Corresponding authors] (PDF)
21. A Serohijos and E.I. Shakhnovich,
“Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics”
(2014), Curr. Opin. Struct. Biol., 26: 84–91. (PDF)
“Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics”
(2014), Curr. Opin. Struct. Biol., 26: 84–91. (PDF)
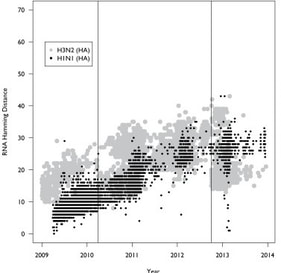
20. E.Y. Klein, A Serohijos, J.M. Choi, E.I. Shakhnovich, and A. Pekoz,
“Influenza A H1N1 Pandemic Strain Evolution – divergence and the potential for antigenic drift variants”
(2014) PLoS One 9(4):e93632. (PDF)
“Influenza A H1N1 Pandemic Strain Evolution – divergence and the potential for antigenic drift variants”
(2014) PLoS One 9(4):e93632. (PDF)
19. A Serohijos and E.I. Shakhnovich,
“Contribution of selection for protein folding stability in shaping the patterns of polymorphisms in coding regions”
(2014) Mol. Biol & Evol., 31(1):165-76. (PDF)
“Contribution of selection for protein folding stability in shaping the patterns of polymorphisms in coding regions”
(2014) Mol. Biol & Evol., 31(1):165-76. (PDF)
2013
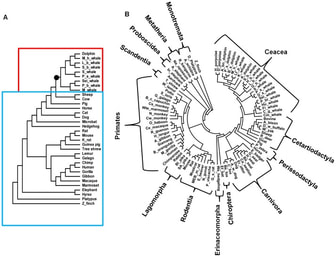
18. P. Dasmeh, A Serohijos, K. Kepp, E.I. Shakhnovich,
“Positively selected sites in cetacean myoglobin increase folding stability”
(2013) PLoS Computational Biology, 9 (3), e1002929. (PDF)
“Positively selected sites in cetacean myoglobin increase folding stability”
(2013) PLoS Computational Biology, 9 (3), e1002929. (PDF)
17. A Serohijos, S.Y.R. Lee, E.I. Shakhnovich,
“Highly abundant proteins favor more stable 3D structures in yeast”
(2013), Biophysical J, 104: L01-L03 (2013) [Cover Article]. (PDF)
“Highly abundant proteins favor more stable 3D structures in yeast”
(2013), Biophysical J, 104: L01-L03 (2013) [Cover Article]. (PDF)
16. S. Bershtein, W. Mu, A Serohijos, E.I. Shakhnovich,
“Protein quality control shapes the fitness effects of mutations in E. coli.”
(2013) Molecular Cell, 49: 1-12. (PDF)
“Protein quality control shapes the fitness effects of mutations in E. coli.”
(2013) Molecular Cell, 49: 1-12. (PDF)
2012
15. A Serohijos, Z. Rimas, E.I. Shakhnovich,
“Protein biophysics explains why highly abundant proteins evolve slowly”
(2012) Cell Reports, 2(2): 249-256. (PDF)
“Protein biophysics explains why highly abundant proteins evolve slowly”
(2012) Cell Reports, 2(2): 249-256. (PDF)
14. M.F. Mabanglo, A Serohijos, C.D. Poulter,
“The Streptomyces-produced antibiotic fosfomycin is a promiscuous substrate of Archaeal isopentenyl phosphate kinase”
(2012) Biochemistry, 51(4): 917-25. (PDF)
“The Streptomyces-produced antibiotic fosfomycin is a promiscuous substrate of Archaeal isopentenyl phosphate kinase”
(2012) Biochemistry, 51(4): 917-25. (PDF)
2011
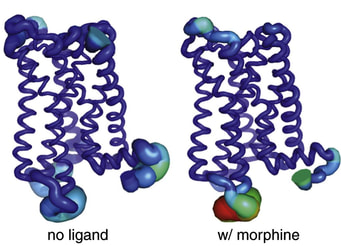
13. A Serohijos, S. Yin, F. Ding, J. Gauthier, D.G. Gibson, W. Maixner, N.V. Dokholyan, L. Diatchenko,
“Structural basis for μ-opioid receptor binding and activation”
(2011) Structure, 19: 1-8. (PDF)
“Structural basis for μ-opioid receptor binding and activation”
(2011) Structure, 19: 1-8. (PDF)
12. D. Tsygankov, A Serohijos, N.V. Dokholyan, and T.C. Elston,
"A physical model reveals the mechanochemistry responsible for dynein’s processive motion"
(2011) Biophysical J, 101(1): 144-50. (PDF)
"A physical model reveals the mechanochemistry responsible for dynein’s processive motion"
(2011) Biophysical J, 101(1): 144-50. (PDF)
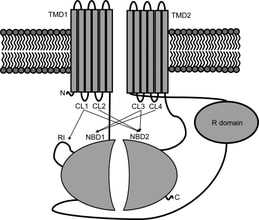
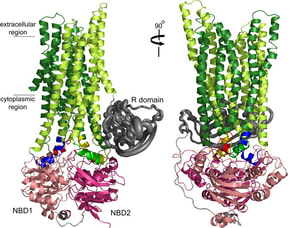
11. A Serohijos, P.H. Thibodeau, and N.V. Dokholyan,
“Molecular modeling tools and approaches for CFTR and cystic fibrosis.”
(2011) Methods Mol Biol. 741: 347-63. (PDF)
“Molecular modeling tools and approaches for CFTR and cystic fibrosis.”
(2011) Methods Mol Biol. 741: 347-63. (PDF)
2009
10. D. Tsygankov, A Serohijos, N.V. Dokholyan, and T.C. Elston,
“Kinetic models for the coordinated stepping of cytoplasmic dynein”
(2009) J. Chem. Phys., 130: 025101. (PDF)
“Kinetic models for the coordinated stepping of cytoplasmic dynein”
(2009) J. Chem. Phys., 130: 025101. (PDF)
9. A Serohijos, D. Tsygankov, S. Liu, T.C. Elston, and N.V. Dokholyan,
“Multiscale approaches for studying energy transduction in dynein”
(2009) Phys. Chem. Chem. Phys. 11:4840-4850. (PDF)
“Multiscale approaches for studying energy transduction in dynein”
(2009) Phys. Chem. Chem. Phys. 11:4840-4850. (PDF)
8. S. Ramachandran, A Serohijos, L. Xu, G. Meissner, and N.V. Dokholyan,
“A structural model of the pore-forming region of the skeletal muscle ryanodine receptor (RyR1)”
(2009) PLoS Computational Biology, 5:e1000367. (PDF)
“A structural model of the pore-forming region of the skeletal muscle ryanodine receptor (RyR1)”
(2009) PLoS Computational Biology, 5:e1000367. (PDF)
2008
7. Y. Chen*, F. Ding*, H. Nie*, A Serohijos*, S. Sharma*, K.C. Wilcox*, S. Yin*, and N.V. Dokholyan*,
"Protein folding: then and now"
(2008) Archives of Biochemistry and Biophysics, 469: 4-19. [*Equal contribution] (PDF)
"Protein folding: then and now"
(2008) Archives of Biochemistry and Biophysics, 469: 4-19. [*Equal contribution] (PDF)
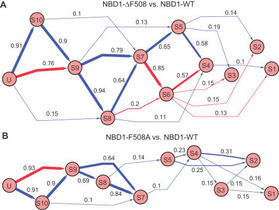
6. A Serohijos*, T. Hegedus*, J. Riordan, and N.V. Dokholyan,
“Diminished self-chaperoning activity of the DF508 mutant CFTR results in protein misfolding”
(2008) PLoS Computational Biology, 4: e1000008. [*Equal contribution] (PDF)
“Diminished self-chaperoning activity of the DF508 mutant CFTR results in protein misfolding”
(2008) PLoS Computational Biology, 4: e1000008. [*Equal contribution] (PDF)
5. T. Hegedus, A Serohijos, N.V. Dokholyan, L. He, J.R. Riordan,
“Computational studies reveal phosphorylation dependent changes in the unstructured R domain of CFTR”
(2008) J. Mol. Biol., 378: 1052-1063. (PDF)
“Computational studies reveal phosphorylation dependent changes in the unstructured R domain of CFTR”
(2008) J. Mol. Biol., 378: 1052-1063. (PDF)
4. J. Hao*, A Serohijos*, G. Newton, G. Tassone, D.C. Sgroi, N.V. Dokholyan, and J.P. Basilion,
“Identification and rational redesign of peptide ligands to CRIP1, a novel biomarker for cancers”
(2008) PLoS Computational Biology, 4: e1000138. [*Equal contribution] (PDF)
“Identification and rational redesign of peptide ligands to CRIP1, a novel biomarker for cancers”
(2008) PLoS Computational Biology, 4: e1000138. [*Equal contribution] (PDF)
3. L. He, A.A. Aleksandrov, A Serohijos, T. Hegedus, L.A. Aleksandrov, L. Cui, N.V. Dokholyan J.R. Riordan,
“Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating”
(2008) J. Biol. Chem., 283: 26383-26390. (PDF)
“Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating”
(2008) J. Biol. Chem., 283: 26383-26390. (PDF)
2. A Serohijos, T. Hegedus, A.A. Aleksandrov, L. He, L. Cui, N.V. Dokholyan, J.R. Riordan, “Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function” (2008) Proc. Natl. Acad. Sci. USA, 105: 3256-3261. (PDF)
2006
1. A Serohijos, Y. Chen, F. Ding, T.C. Elston, and N.V. Dokholyan, “A structural model reveals energy transduction in dynein” (2006) Proc. Natl. Acad. Sci. USA, 103: 18540-18545. (PDF)
Patents
|
“Mu-opioid receptor binding compounds” L Diatchenko, W Maixner, NV Dokholyan, F Ding, A Serohijos, S Yin, International Patent Application PCT/US12/40168 May 31, 2012; U.S. Provisional Patent Application No. 61/491,828 filed May 31, 2011
|